Tutorial collection
A step-by-step tutorial explaining how to use SUBA is available here:
SUBA5 Search Help ¶
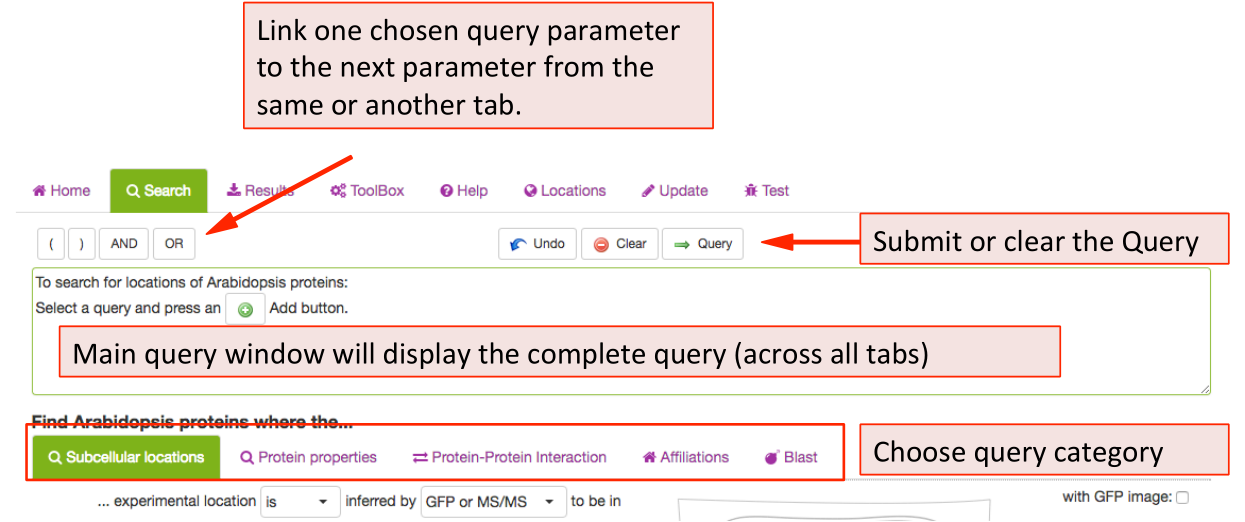
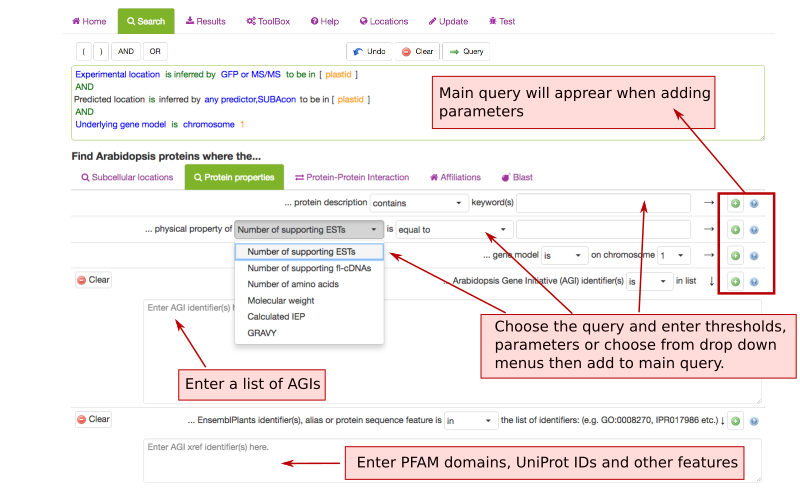
When clicking on the Search function the tab opens the query builder menu. In SUBA5 there are more options and categories of queries you can use to interrogate the SUBA data set. In order to enable the user to find the desired search parameters we have introduced search categories. Each category is stored under a tab. The user can choose a parameter from any tab and add it to the query. The query will appear in the query window at the bottom after clicking button. Different search categories can be combined using the AND/OR buttons in between parameters above the query window.
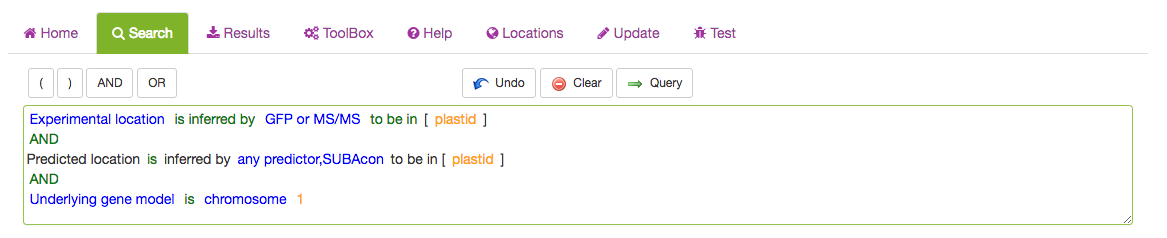
The full query can be seen in the query window. For details about each category please see search category sections in the tutorial below.
Subcellular location Search Tab ¶
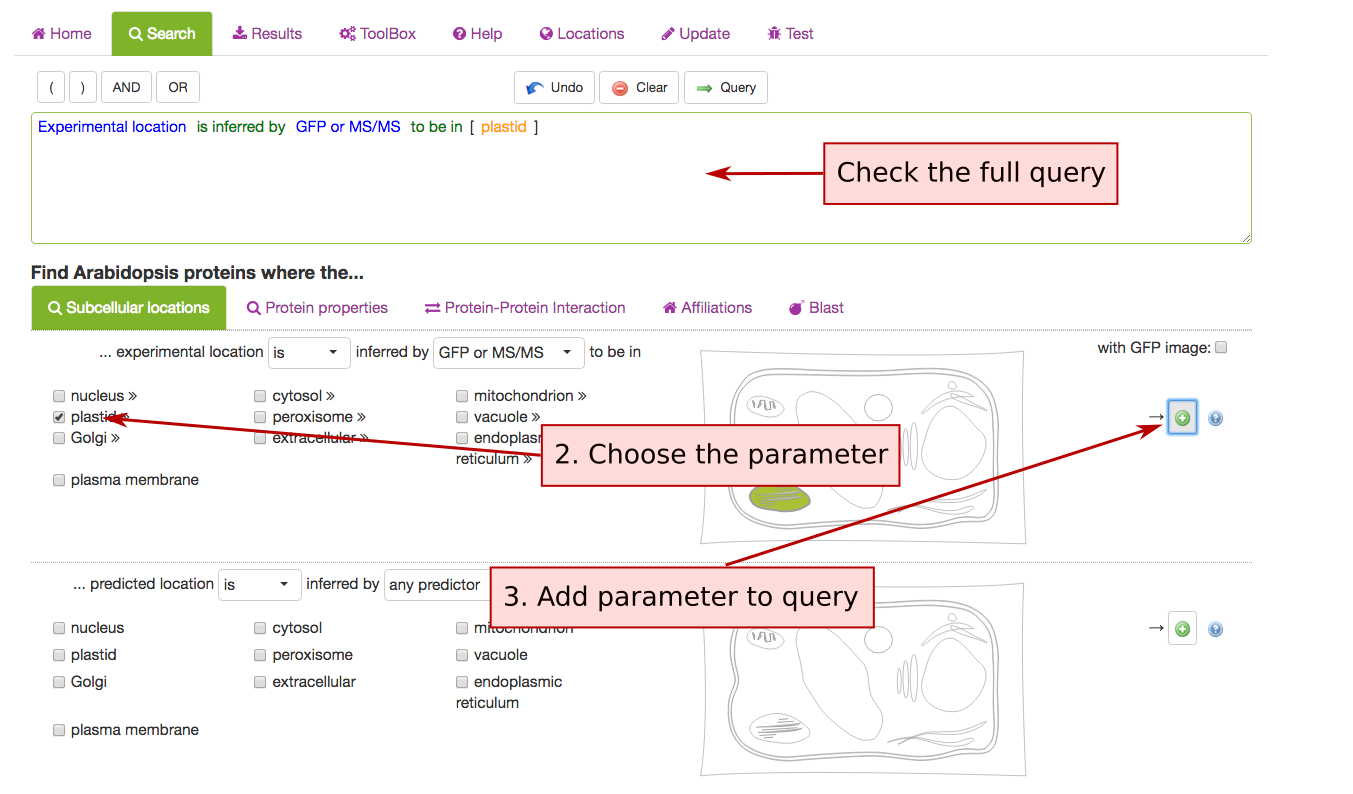
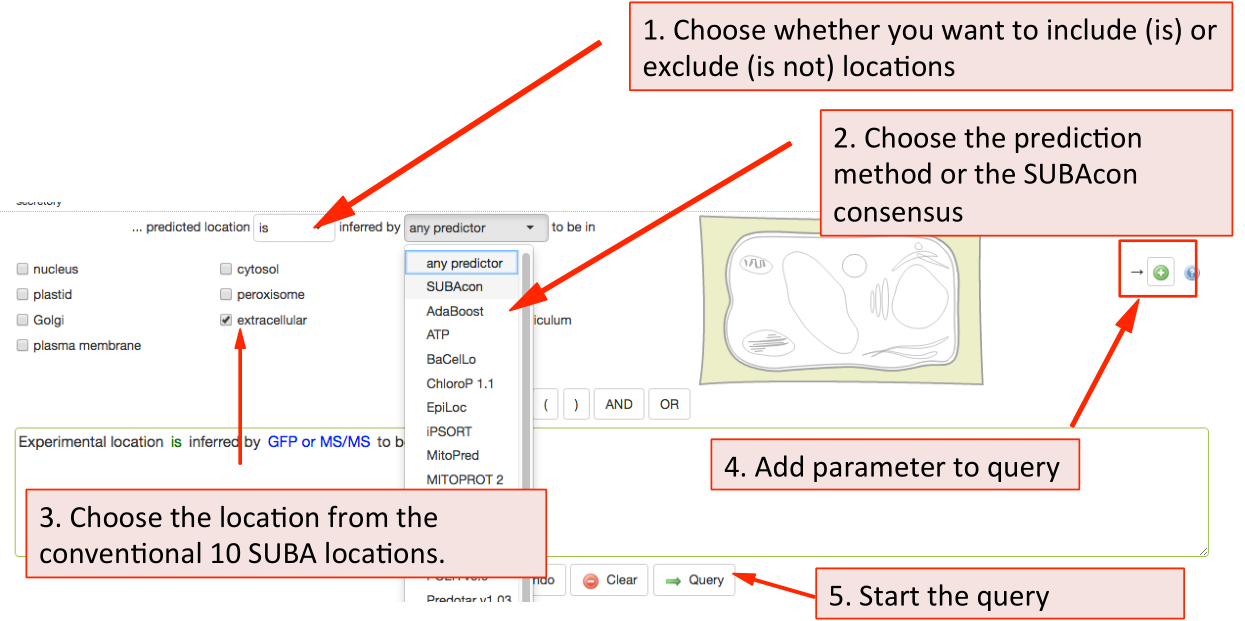
This tab contains queries for limiting proteins based on their localisations. SUBA5 has 2 main categories of localisations. You can search for experimental localisations, which is the top query parameter.
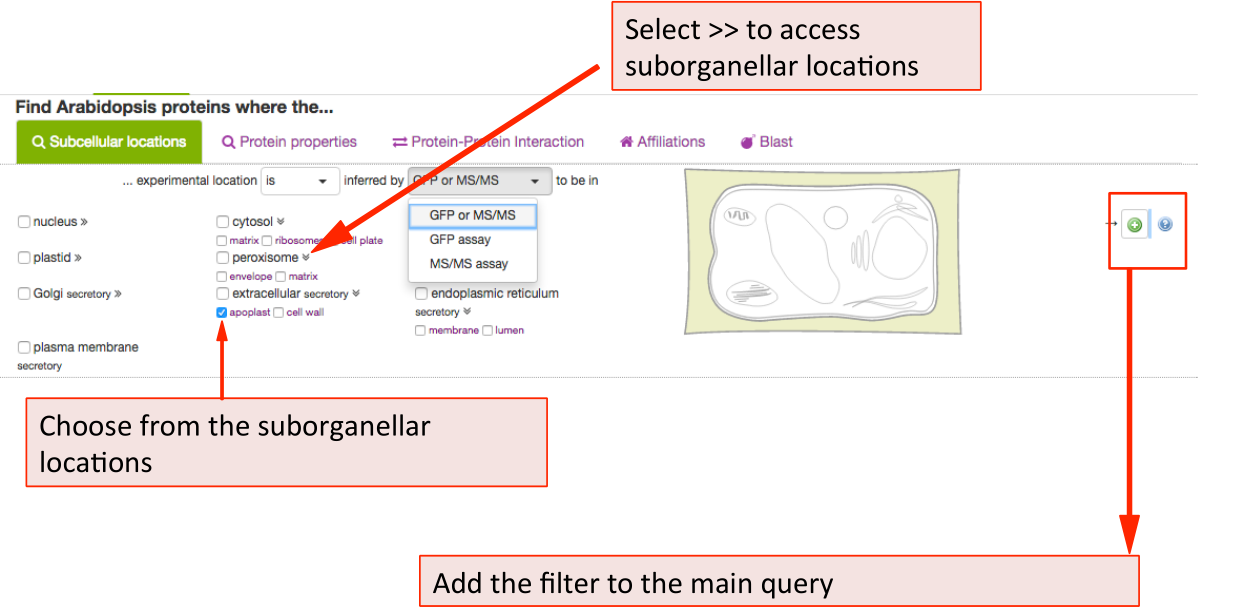
The parameters such as in/exclusion of particular compartments and methodology can be chosen from the drop down lists. For choosing a subcellular location, tick any of the box or structures in the cell schematic for the conventional SUBA location categories. For expanded suborganellar categories, click on the >> to expand the list.
This will maximise the localisation view. Click on the desired location. For choosing more then one location, keep ticking more boxes. When choosing the whole compartments
(extracellular), this will automatically include the suborganellar locations (apoplast, cell wall). For
only searching for apoplast, untick extracellular and only tick apoplast. Then add your parameter to the query by clicking the  button.
button.
Similarly, to filter for prediction data choose the inclusion and exclusion and the type of predictor from the drop down list. Through this search option, you can also filter by our consensus call output when choosing SUBAcon.
Once you have added all desired parameters to the query window you can check your query and submit it using the Query button. Your retrieved results will be automatically displayed in the Results tab when ready.
Protein Properties Search Parameters ¶
The Protein property tab lets you filter SUBA data for protein annotations, physical properties and chromosomal locations. This tab also contains the option to enter a list of AGIs or text containing AGIs. A new query in SUBA5 lets you also filter for protein aliases, PFAM domains, EC numbers, pathway annotations, structural features and other annotations.
Protein-Protein Interaction Search Parameters ¶
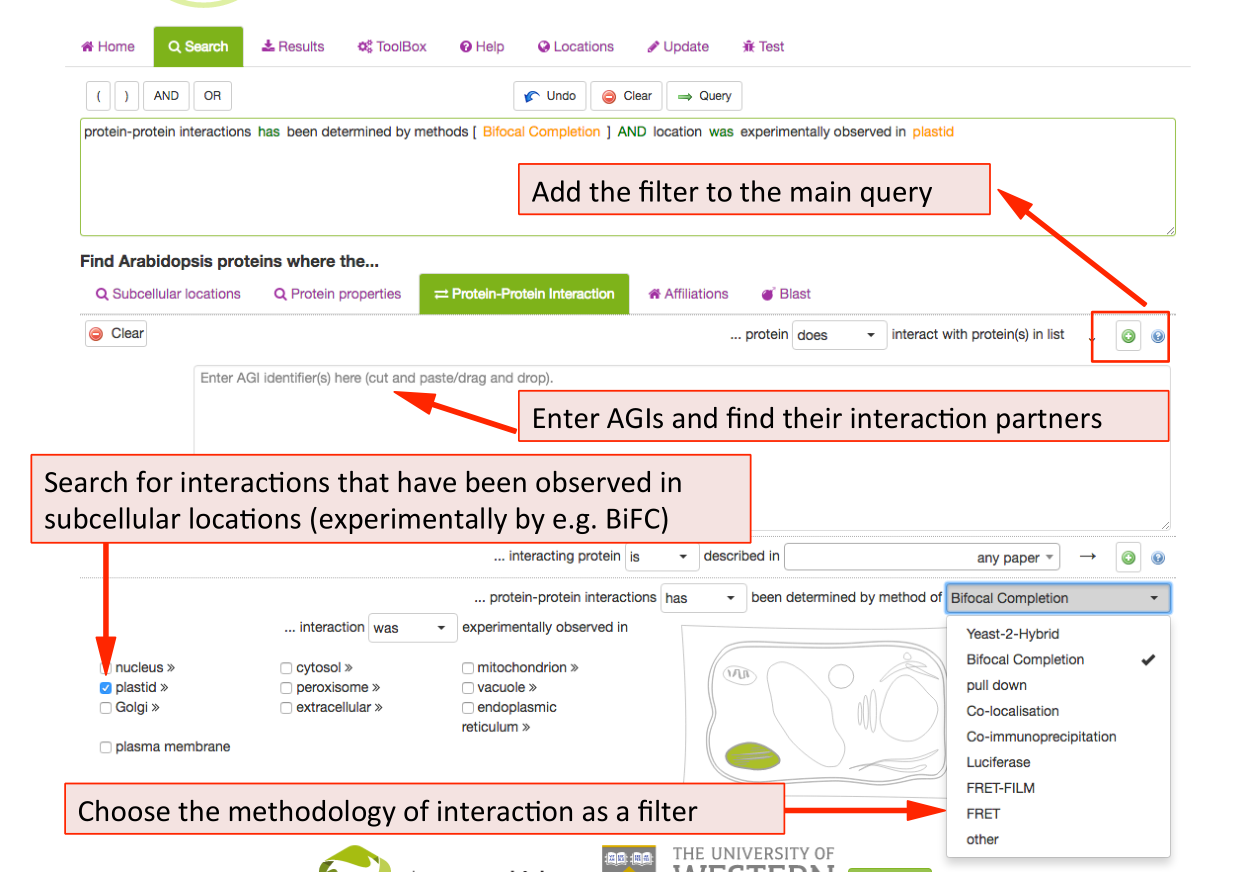
New in SUBA5: In addition to protein‐protein interactions (PPI), there are now experimental localisations from observed protein-protein interactions (PPI) such as Bifocal completion Experimentation. The PPI search tab was included to provide a straightforward access to a number of PPI queries. Besides the conventional search for existing PPI partners by entering AGIs, SUBA5 users can now discover PPI proteins that have been experimentally shown to interact in a particular compartment. At the same time, the drop down menu also allows for the choice of PPI methodology. Other search options for PPI data include the isolation of PPI studies.
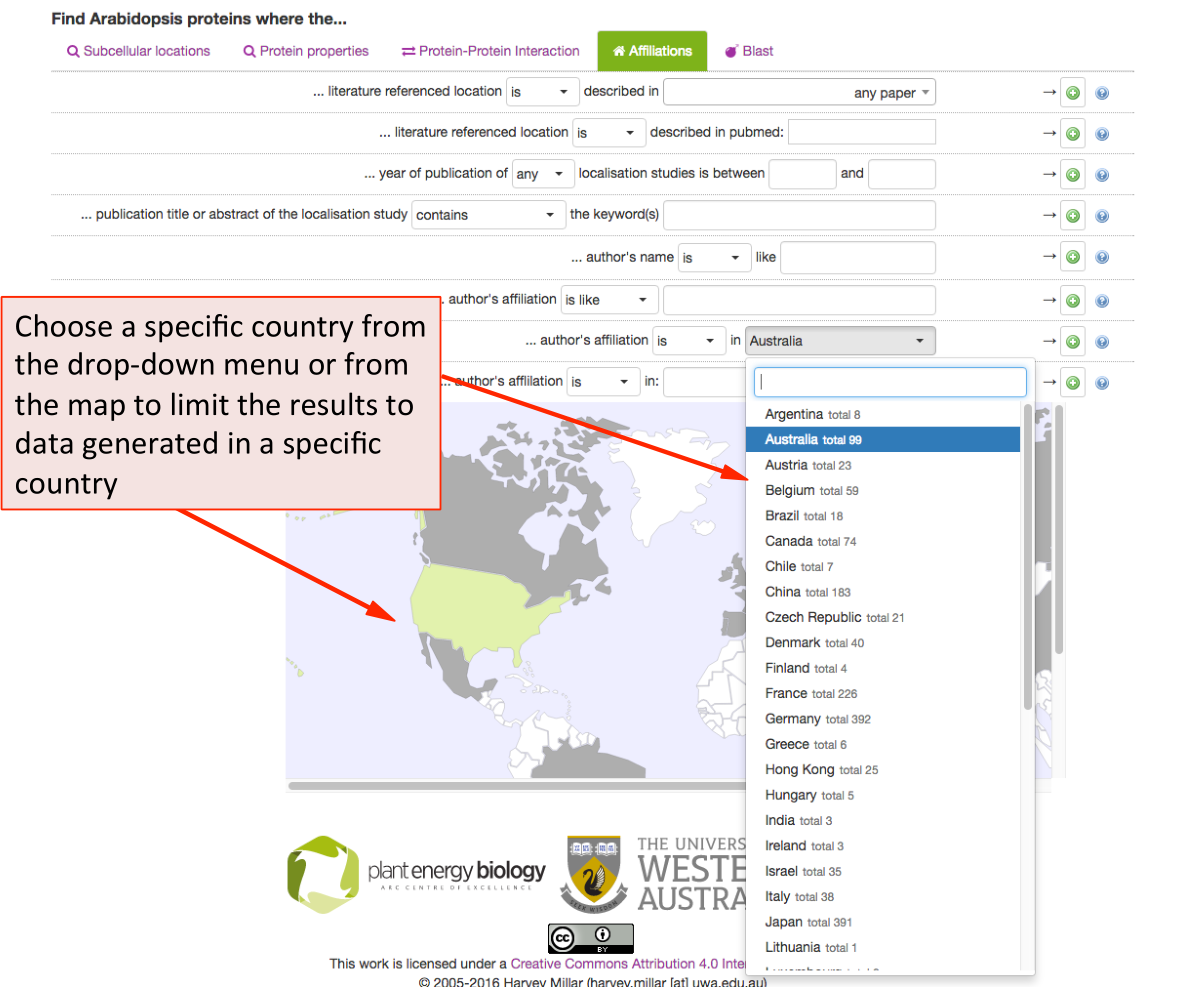
Affiliations Search Parameters ¶
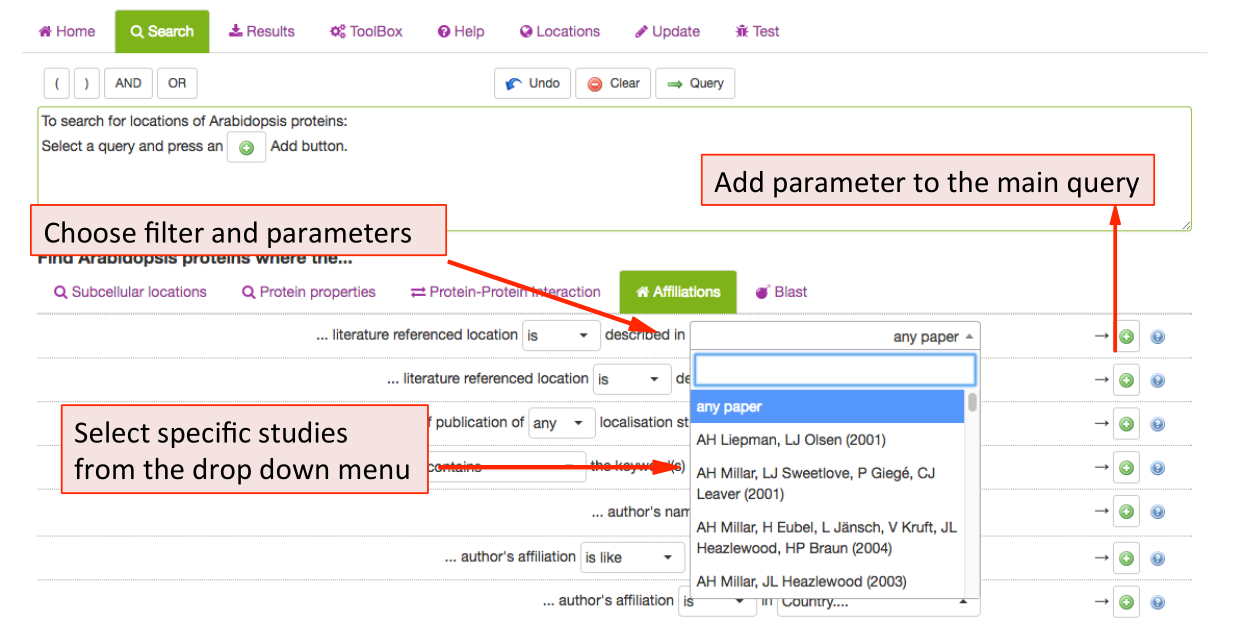
This tab allows SUBA5 users to find experimental localisation data from specific
authors, institutions, countries or filter by year of publications. To limit
the results a particular study, choose from the publication list in the drop-down menu. The results can also be filtered by any author (not just first author) as well as by year or range of years of publication. For adding any of the parameters to the main query press the  button.
button.
SUBA5 allows the search for data by country of origin of the experimental study. Using the drop‐down menu shows the countries and number of studies that have contributed to the SUBA5 data set. When using the map for choosing a country, the grey countries indicate a contribution to SUBA5 whereas white countries have not contributed data sets to SUBA5. Green indicates a chosen country.
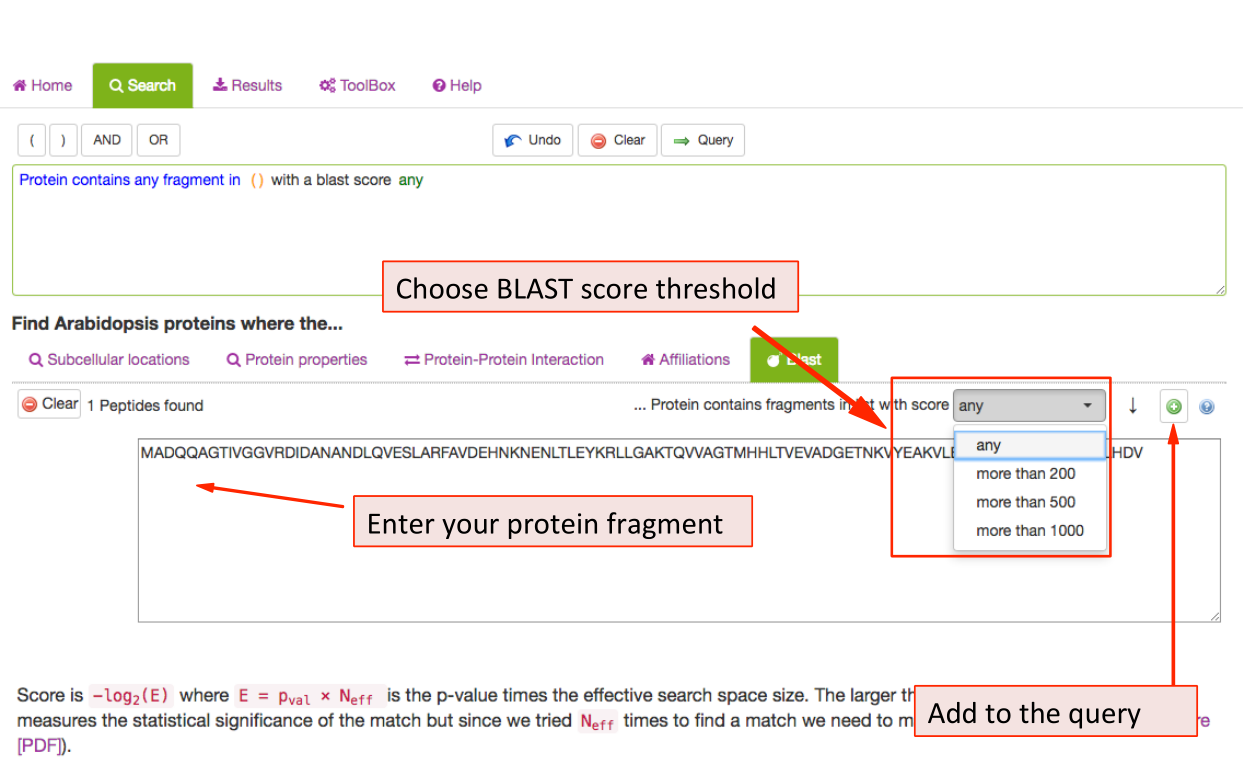
BLAST Search Parameters ¶
The BLAST tab contains the BLAST tool equal to the one in the BLAST panel labelled ‘Find your closest AGI!’ on the SUBA5 homepage. The user can enter a sequence and retrieve data from Arabidopsis proteins with sequence similarity. The results can be filtered using the BLAST score as a threshold. The score measures sequence similarity in respect to sequence length. The BLAST hit AGIs are retrieved and the data for the AGI linked protein is retrieved from SUBA5.
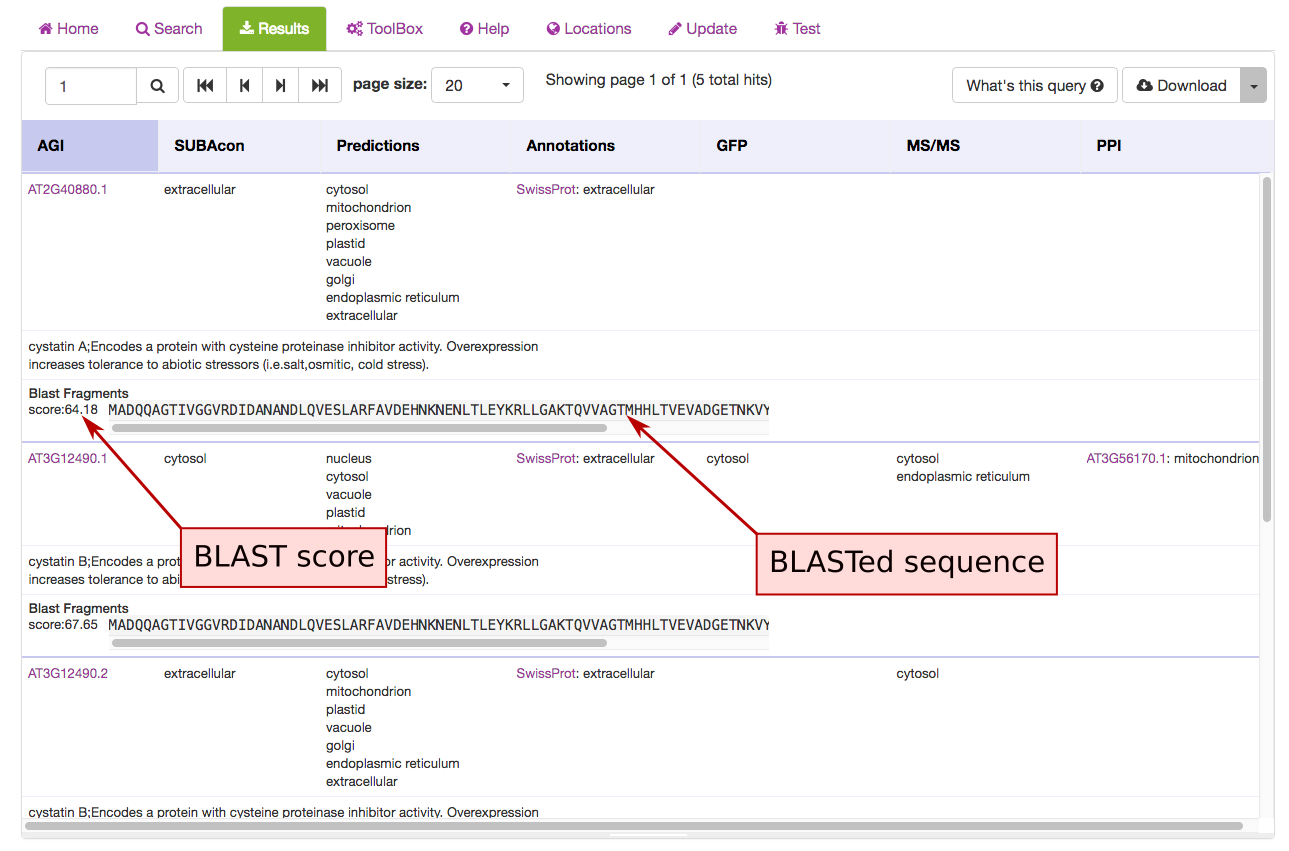
The hits are displayed in the results view and each hit shows the BLAST score and aligned protein sequence.
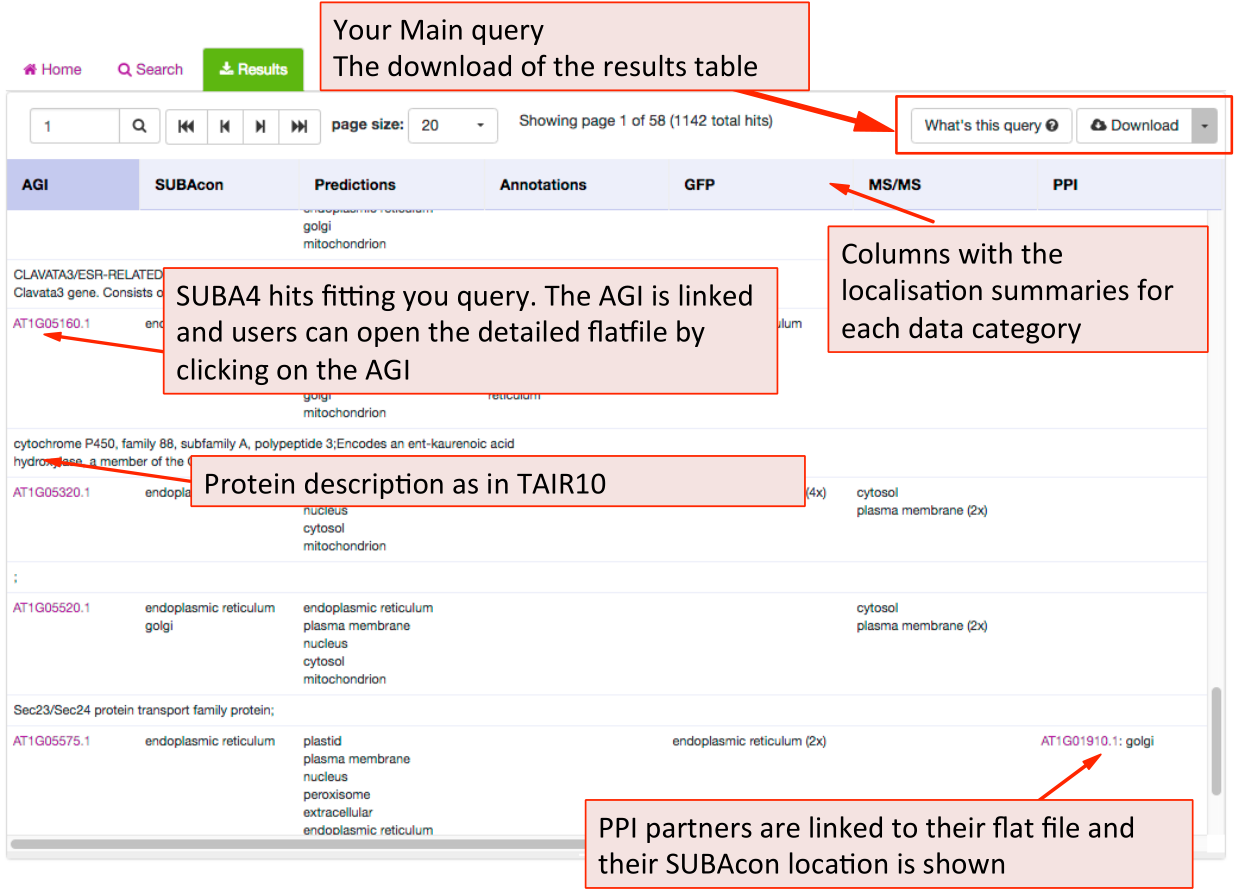
SUBA5 results tab ¶
The results tab will automatically be activated when the query is submitted. SUBA5 users will be able to see the query by clicking on the “What’s this query” button in the top left. The results can be downloaded as a table format using the download button.
The results are presented in table format. The columns can be customized towards the preference of the user. The first column shows the AGI of the proteins fitting the submitted query and the description for the protein below. This is followed by the consensus call derived from SUBAcon. Each of the individual localisation data columns show the summary of the data for the category. For a more details view for each category the user can access the factsheet by clicking on the AGI.